The aldehyde dehydrogenases (ALDHs) are key enzymes which play a vital role in many growth and developmental processes. Adverse environmental stresses such as drought, high salinity and low temperatures badly affected the plants as they are immobile. These stress conditions often eventuate in the high accumulation of reactive oxygen species (ROS) which lead to oxidative stress and cell damage.
Yergaliyey et al. (2016) stated that, “oxidative stress is coupled to the oxidative degradation of lipid membranes, which could generate a number of aldehydes”. It has been reported that too much accumulation of aldehydes interfere with metabolites and become toxic. Aldehyde dehydrogenases (ALDHs) is a protective proteins which plays a significant role in the metabolism of endogenous and exogenous aldehydes in order to cope up with the cellular damage of aldehydes and mitigate oxidative stress in plants.
ALDHs are reportedly present throughout all taxa in both eukaryotes and prokaryotes. ALDHs are divided in to 24 distinct families based on protein sequence identities. Koncitikova et al. (2015) notified that, out of 24, 13 ALDH families have been well characterized in plants.
Cao et al. (2021) mapped out a research that was aimed to identify and analyze ALDH gene family in genome of Eucalyptus grandis, an important woody tree. Study was conducted during October, 2018-2019. The in silico analyses were carried out at Hung Vuong University, Agricultural Genetics Institute, Lebanese University, Hanoi Pedagogical University 2, and Hanoi National University of Education.
The ALDH family members in the E. grandis genome were identified by a basic local alignment search tool (BLASTP) against the E. grandis proteome database using known Arabidopsis and Vitis ALDHs as queries. Sequences were analyzed by various bioinformatics tools.
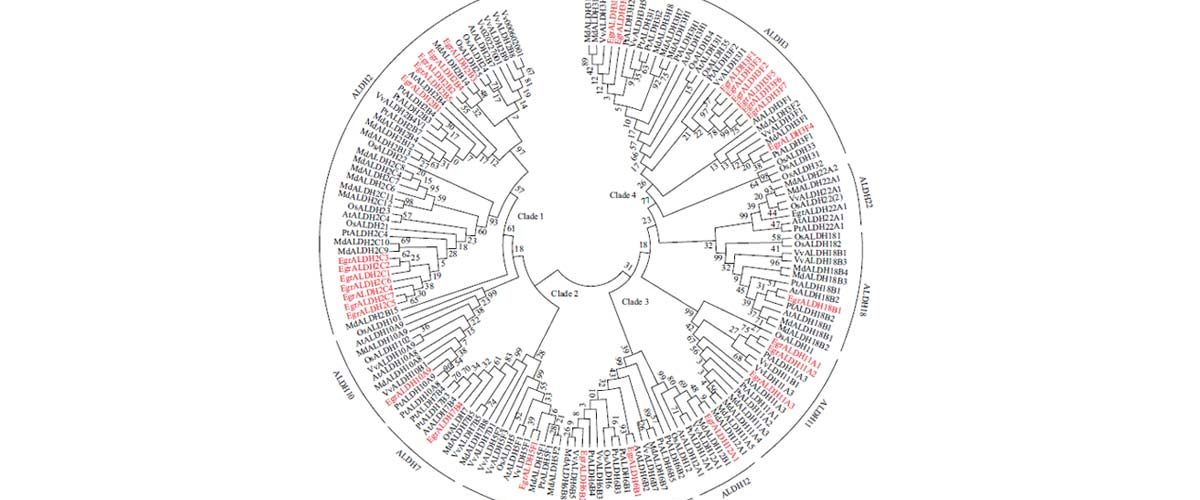
The results of this study exhibited that a total of 32 members of EgrALDH gene superfamily were identified in E. grandis and their characteristics including chromosomal distribution, sub cellular localization, protein features, gene structure and phylogenetic tree were subsequently analyzed. It has been predicted that the segmental and tandem duplication events (20 out of 21 duplicated pairs) might be the major mechanism of the expansion of EgrALDH genes. The EgrALDH genes have differentially expressed in young leaves, mature leaves, shoot tips, phloem, immature xylem and xylem.
A total of 32 EgrALDH genes, particularly 12 EgrALDH2s, nine EgrALDH3s, one EgrALDH5, two EgrALDH6s, one EgrALDH7, one EgrALDH10, three EgrALDH11s, one EgrALDH12, one EgrALDH18 and one EgrALDH22 identified inthe E. grandis genome and their general features were provided. Using various in silico analysis, this study provided the first sight into the understanding of the EgrALDH superfamily, including the protein characteristics, exon/intron organization, gene location, phylogenetic tree. This study demonstrated that the duplication events, including segmental and tandem duplication, might play the main roles in the EgrALDH genes.
In conclusion, this study will be helpful for researchers as it act as a substratum of the EgrALDH gene superfamily in E. grandis for further functional characterization.
Keywords:
Genome-wide Analysis, Aldehyde Dehydrogenase (ALDH), Eucalyptus grandis, Bioinformatics Methods, oxidative stress, reactive oxygen species (ROS).














Add comment